How we can extend the limits of DNA ethnicity estimates to make these tests more reliable and relevant for more people? A look ahead at the next generation of DNA ethnicity estimates.
This is the final article in Jayne’s excellent, in-depth series on Ethnicity Estimation. Many ancestry testing clients have felt confused by their results, perhaps especially when they receive ethnicity estimates that differ between companies or version upgrades. Take a look back at previous posts to get a broader picture of the nuts and bolts of the scientific process, and different tools that you can use to confidently evaluate your results in hand. You may find just a little bit of awe as well that all of this can be determined from a vial full of spit. That’s pretty incredible!
Recently, I wrote about the current limits of DNA ethnicity estimation. A natural follow-up question practically screams for an answer: how we can extend the limits of DNA ethnicity estimates to make these tests more reliable and relevant for more people?
The good news is that the DNA testing companies want this, too, and are working hard to achieve it. In fact, your DNA ethnicity results may have (even recently) been updated to more refined results, as companies continue to expand their reference panels and refine the complex math used to calculate your ethnicity pie chart.
Whether you find these updates informative or perplexing, they do represent significant, state-of-the-art advances. And many more improvements are coming. Some are currently underway, and many are still in the concept stage, but will be rolled out as they are ready.
Improved SNP selection, relevant to more world populations
A significant next step underway is to make ethnicity tests relevant to more people with roots in all world populations. When this technology was in the initial development stages, the first large databases were heavily weighted with representatives from Europe. Since then many companies have made it a focus to significantly expand their reference panels. 23andMe recently announced some initial findings from its African Genetics Project. About 50,000 participants living in the United States with four grandparents having roots in the same African ethnic group were recruited for this large effort.
Earlier in 2019 AncestryDNA announced an expansion of 94 new DNA communities to serve clients of African descent in the Americas and Caribbean. Along with a continually expanding sample base for Asian and Native American clients, the representation of diverse world populations in these databases is steadily improving.
With this expanded ethnic representation comes a need to reexamine the 700,000 SNPs that had previously been selected for use with primarily European populations. SNPs are generally not equally useful for all world populations. A SNP by definition is a single base position on a chromosome that takes usually one of two base letter states. In this case, an individual inherited a T from his father, and a C from his mother in the same position on the chromosome.

For a SNP to be useful for studying a certain population, it has to vary in that population. For instance if everyone from Korea has a T at a particular SNP, this isn’t a marker that will helpful to demonstrate genetic differences within Korea. It would be much more informative to use a marker where perhaps 30% of Korean individuals showed a C allele, and 70% have a T. This is a SNP that has a lot more to say about the genetic history of Korea than a SNP where everyone is the same.
All companies have gone through more than one version of the SNP panel they use. Through these revisions and other studies, the scientific community has identified which SNPs are more informative for different world populations. As a result the companies are now better able to hand-select SNP markers that are targeted for maximum usefulness in an ethnically diverse setting. These newer SNP panels have been in use at the companies for the last couple of years. Especially if you originally tested before this time frame, it is worthwhile to examine your current report or contact your testing company to understand whether your personal results were produced using their most recent SNP panel. Customers with non-European roots, in particular, should see improved ethnicity estimation results that better reflect their specific genetic heritage.
Greater granularity from different types of genetic markers
SNPs are currently the fastest and cheapest way to generate a broad survey of an individual’s entire genome. About 700,000 points, scattered across all of the chromosomes, give a pretty good snapshot of a client’s DNA. But there are other types of markers that provide a different angle on the history of an individual genome. STRs (Short Tandem Repeats) were heavily used for many years, but fell to the back burner with the mass discovery and mainstreaming of SNPs. STRs have a lot to say about an individual’s ancestry, with the potential to shed light on more recent heritage than SNPs.
Another genetic test, Whole Genome Sequencing (WGS), is a process that is still too expensive for the mass market but captures the maximum detail of an individual’s genome. Since it reads each point of the 3 billion base sequence of As, Ts, Cs, and Gs in order across all chromosomes, you can’t get a more exhaustive look at the genome than that! As the WGS process is streamlined, prices will continue to decline and we can expect to see this option made more accessible to the general public. STRs and WGS can both play a role in providing different detail than we currently see in ancestry testing.
STRs and SNPs
In the era following O.J. Simpson’s white Bronco, the power of STRs in identifying humans from their DNA hit center stage. STRs are short sequences of DNA that have a repeating, stuttering pattern for several units looking something like this:
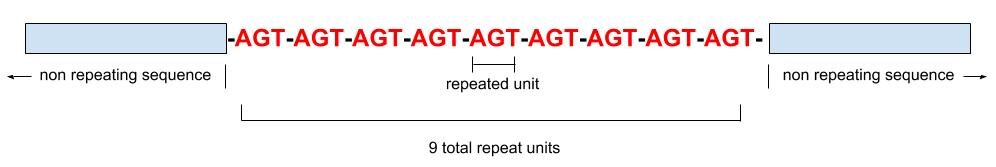
STRs are reported as the total number of repeated units– 9 repeat units are shown in the example above. The segments of DNA that contain repeats are inherited at each generation, so family members will often share the same number of repeats for the same STR marker. STRs are much more prone to mutation than SNPs (SNPs are basically considered a one-time event in the entire human pedigree). The molecular machinery that replicates DNA is prone to slippage in these repeated regions, so the number of repeats can mutate over the generations, either to have more repeat units or fewer. STRs are also prone to back mutations, which means they don’t necessarily keep getting longer or shorter according to a previous trend, but can go up and then back down in number of repeats in any given generation.
Using many individual STRs together in combination is a powerful way to link individuals to close families. When your scope expands to the entire world though, there are some challenges to using STRs for determining an individual’s ethnicity. Using STRs to generate a DNA fingerprint for a region is possible, but it would not be unexpected to see portions of that DNA signature pop up in other places around the world in populations that are not closely related. This happens because STRs mutate relatively quickly, and groups that are not closely related can randomly experience the same STR states. This situation is called parallel mutation, or Identity-By-State (IBS) rather than Identity-By-Descent (IBD) where DNA signatures are in common due to recent inheritance on a time scale that is relevant for genealogical questions. The presence of IBS mutations makes it difficult to interpret large-scale STR data from diverse groups.
As the use of SNPs was becoming more widespread in the early 2000s, researchers began to use them in tandem with STRs to mitigate the effect of IBS mutations, and still access the finer-scale granularity that is available with STR markers. Most of this research happened in the Y-chromosome, which was a great place to begin to apply these systems without the added complexity of autosomal recombination.
The way that this played out in large population studies was to first generate a detailed SNP profile for each study participant. This allowed researchers to separate individuals into pools of independent SNP-defined groups, and then they overlaid STR data on top of that. Even though IBS STR data might show up across the world due to the frequent mutation rate, they were able to keep individuals separated into more closely related, genealogically relevant groups because of their defined SNP profiles. And then they used the STR data to draw out finer-scale relationships within those larger SNP-defined populations, detecting ethnicity and family groupings with greater resolution than with SNPs alone.
Computational models have progressed enough today to apply a similar process to reap the power of combined autosomal SNP and STR data. In the autosomal genetic system, at every generation there is a reshuffling of each chromosome as sections of the mother’s and father’s DNA are recombined and mixed together.
Phasing is the process of determining which section of autosomal DNA came from each parent. This is now a process that is well-defined and is regularly practiced with the autosomal SNP data that is used in ethnicity tests. Phasing STR states could also be accomplished with similar methods, predicting which STR allele came from which parent, and eventually being able to trace STR states back through pedigrees to larger ethnicity groupings. By overlaying phased STR data onto more deeply defined SNP profiles, confounding IBS mutations could be separated out into more meaningful closely related groups. The power of these combined markers could give way to more detailed profiles for genetic ethnicity than is currently available with SNPs alone.
The timeframe of resolution for the lens of ethnicity detection offered by SNPs is not well defined, but are mostly suggested as being sensitive to the last several hundred to one thousand years. By nature of their more aggressive mutation rate, STRs shorten that lens by linking individuals and groups that are more closely related in more recent generations. The downside is that producing STR data is slower and more labor intensive than SNP analysis.
Whole Genome Sequencing
If applying STR data is seen as an intermediary step to obtaining greater genetic resolution for ethnicity detection, whole genome sequencing (WGS) is the ultimate granular lens for viewing what your DNA has to say.
It took 13 years and $2.7 billion to sequence the first human genome. 17 years later, today it takes about 2 days and costs $1,000 for a good high-coverage scan. It used to be news when a genome from another individual was sequenced. Subjects from diverse ethnicities were selected for sequencing and entire scientific papers were published comparing the differences in the new ethnic sequence to the original data from a European male. One such study from 2014 sequenced an individual from Turkey and found 3.5 million SNPs in the 3 billion base pair sequence. 480,396 of those SNPs were novel to popular SNP panels being used at the time. This and other studies like it highlight the presence of ascertainment bias in the selection of SNPs that are widely used in the ancestry testing industry.
Ascertainment bias occurs when population sampling is not random (i.e., heavily weighted toward Europe) and SNP discovery favors SNPs with certain characteristics (highly informative for Europe but not others, SNPs that are ancient and present in much of the world population and therefore not as distinguishing, or preferring SNPs in certain genomic regions over others, for example). As we spoke of earlier, there have been concerted efforts to expand the usefulness of SNP panels to many more world populations, but it’s still possible with only 700,000 points of a much larger genome to miss capturing the full richness of worldwide population diversity.
One of the ideas with WGS is that it’s not necessary to do special subpopulation studies to select SNPs that ensure that the massive diversity present in the world is represented. All of the variation is captured at the outset when the entire genome is sequenced, so ascertainment bias is dramatically reduced or eliminated altogether.
Discovering private variants that exist in under-characterized regions of the world will reveal finer-scale relationships between individuals, families and populations. And likewise in areas that are already well represented, the maximum granularity represented with WGS will enhance the resolving power of ethnicity and relationship tests that are currently performed with SNPs.
A quick internet surf will show you many ways you can obtain your own WGS by the end of this week if you want. That’s pretty cool, right? Black Friday sales have been known to reduce the $1,000 price tag to as low as $199. The trouble with this is that the interpretation tools for drawing meaning out of your WGS are not yet developed. You’ll mostly just get the satisfaction of carrying around two thumb drives full of your intimate genetic information. But as the price of WGS is driven down, this will be the ultimate direction of ancestry genetics. So we wait patiently. A lot has happened over the last 17 years since the first human genome was sequenced. What will the next 17 hold? I don’t know if we can even conceive it.
 Interested in more insight on what your ethnicity results mean? Check out our free guide on DNA ethnicity estimates.
Interested in more insight on what your ethnicity results mean? Check out our free guide on DNA ethnicity estimates.
Have you read Jayne’s entire ethnicity estimation series? It’s been one of the most popular things on our website. Read it from the beginning. And thank you for sharing them with your friends who want to better understand their DNA test results!


