You’ll feel so much smarter about your varying DNA ethnicity results after you read this article! Here’s why you should think about them more as “historical communities.”
This is the second article in Jayne Ekin’s series on taking a closer look at DNA ethnicity. Read the first article here.
Humans have been traipsing around the earth with their DNA in tow for a long time. When you combine “DNA” and “a long time,” you end up with a wealth of astonishing variation that we are the inheritors of today: Mongols with green eyes and blond hair in the grassy plains of western Asia, singular shovel-shaped teeth that are found only in East Asians and Native Americans, and Bantu-speaking black South Africans possessing a distinctive Semitic DNA signature, to name just a few. The accumulated DNA mutations of thousands and thousands of years, along with our global wanderings, has gifted the 7.7 billion people on the planet today with a gorgeous amount of diversity.
Our DNA is our history
It’s no surprise that this diversity tends to show up systematically, often linked to a geographic or cultural heritage. Our physical traits link us not just to our immediate relatives, but also to the larger communities and societies where our ancestors lived. Until very recently we couldn’t “see” our DNA the way we can see physical traits like skin tone, hair type or freckling. But our DNA has been leaving breadcrumbs along the trail this whole time, linking us back to our historical communities much the same way our physical characteristics do.
The unraveling of the genetic systems within our DNA has rapidly accelerated over the last 20 years. Today most ancestry clients are tapping into their genetic code with autosomal tests that examine the 22 chromosomes that are inherited from both mom and dad. Despite the complex reshuffling process these chromosomes are subjected to each time a new human is created, science has recently made remarkable strides decoding the rich ancestry information contained therein.
Y DNA shows our human family tree
A separate genetic system, the Y-chromosome, has already given us an abundant picture of human history and was riddled out much earlier due to its straightforward inheritance pattern: strictly father-to-son without any input from mom. Although the autosomes and Y-chromosome contain different information, their distinct lenses on human history compliment and corroborate one another. It turns out that the Y-chromosome has a lot to say about where our ancestors have been keeping house, and how long they’ve been at it.
SNPs (single nucleotide polymorphisms) are a genetic mutation of a single base at a particular position along the genome, the same type of genetic marker used in consumer autosomal tests, that also occur on the Y-chromosome. They represent a single mutation event that happened in a person somewhere along the line of the great human pedigree. Once the mutation was acquired it was passed down to all of that person’s descendants. New SNP mutations happened by chance in some of these descendants, who in turn passed it on to succeeding generations, along with any they had inherited from their predecessors. With all of the generations that have passed since the early days of the human family to now, each of us are the privileged bearers of a menagerie of mutations that reflect our personal ancestry and also show us how we are linked to the great human pedigree.
In the Y-chromosome, scientists have catalogued the geographies where each of these SNP mutations are seen in high frequency in the world, and have also derived when these mutations occurred. What emerges is a tree of the human family that identifies the most ancestral genetic state that forms the trunk of the tree, and subsequent Y-SNP mutations that have occurred along the way forming many branches off the trunk. This tree describes a history in which generations of intrepid humans were populating new parts of the world, and time estimates of when they were going about it.
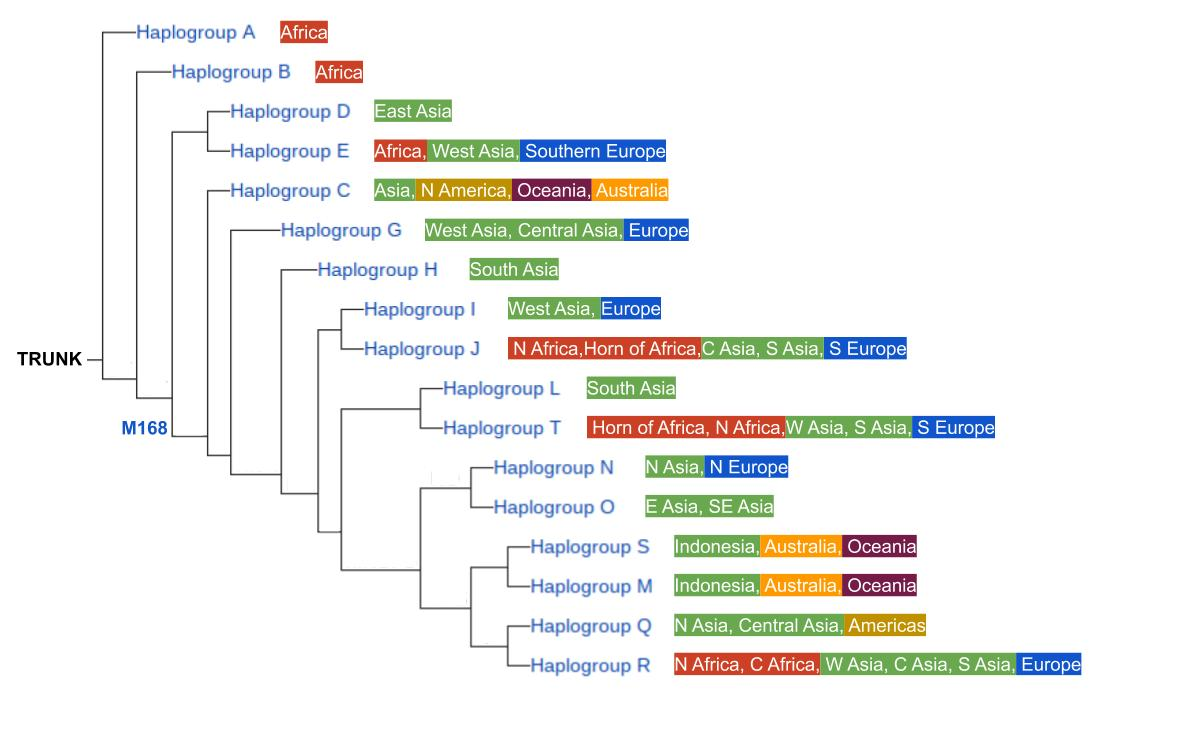
This tree is simplified and generalized for space and clarity, but shows the major haplogroups that form the largest branches of the Y-chromosome SNP tree. The genetic state that defines Haplogroup A is ancestral to all Y-chromosomes found in humans today, and the subsequent mutation that created Haplogroup B became the first major branching event in genetic history. A significant mutation event downstream, called M168, gave rise to all subsequent branches (Haplogroups C through R). Excluding those in Haplogroup A and B, every male in the human family possesses the M168 mutation along with any other mutations their ancestors accumulated along the way that ultimately places them in a different and younger branch of the tree. Individuals that possess only the Haplogroup A and B genetic states are found only among groups of native Africans. These are such distinctive genetic characteristics that they are not found elsewhere in the world. Further, there are geographic characteristics associated with each subsequent haplogroup, which gives a sense of how humans have been spreading and sharing their genes over these many, many generations.
DNA ethnicity goes waaaay back
How many generations? Way back. Way, way, waaaaay back. The M168 SNP, among the eldest of ancestral mutations, has an estimated age of 48,000 years. That’s a lot of years that our DNA has been forging new generations, developing new SNP mutations, and leaving breadcrumbs around the world. As you move out into the newer branches of the Y-SNP tree the ages of founding mutations are more recent, but still go way back. The mutation that defines haplogroup Q is estimated to have occurred 24,500 years ago, the haplogroup R mutation, 27,000 years ago. Still certifiably ancient.
DNA didn’t stop mutating when the major haplogroup branches were created. There are entire sub-tree structures that have been teased out of each major branch defined by newer SNP mutations that form increasingly specific lineages within the large super-groupings. As you move out into the twigs and leaves of the tree, some SNP mutations are brand-spanking new. J2b-M205 is a small sub division of Haplogroup J found in Italy, Greece and Northern Turkey that clocks in with an age of 4,000 years– just a baby by comparison.
It’s certainly interesting to dive deeply into the Y-chromosome, but ethnicity estimations today are based on autosomal DNA, right? While these are separate genetic systems, patterns detected in the Y-chromosome give us some clues into what the whole human family has been up to over the ages, not just males—after all, males didn’t create new generations on their own. And just as we see SNP mutations accumulating and creating branches and sub-branches over time in the Y-chromosome, this is also the order of things in the autosomes. While in the autosomes there is the substantial added complexity of generational recombination with the mixing of separate genetic lineages from mom and dad, it still mirrors patterns seen in the Y-chromosome history–larger branches carrying ancestral mutations, with newer mutations accumulating over time reflecting more recent population groupings.
As scientists have developed algorithms that predict ethnicity for individuals, they have observed that the most confident predictions are observed at the continental level–the level where major branchings of the human family tree occur. The algorithms will produce estimations for sub-regional and local geographies, but they bear substantially less confidence than at the continental-level. As seen in the Y-SNP tree, the major branches of the complex autosomal tree follow continental patterns and are defined by groups of mutations that are quite ancient. More recent SNP mutations are certainly present out in smaller sub-branches, but these are notoriously difficult for algorithms to sort out due to substantial sharing of DNA among recently and closely related populations.
Autosomal DNA ethnicity results are messy
Some ancestry DNA clients have wondered in the face of conflicting ethnicity results how it is for example that they can display a marker for Tuscany in one test, but it shows up as Calabria in another. How can I have both a Tuscan and Calabrian marker? One of the answers to this is that there are no Tuscan or Calabrian markers. These are population groupings that algorithms are attempting to make out in the leafy twigs of the recent human family tree, far out from the mutations that define the sturdy major branches. Under some conditions there are some recent communities that will develop distinct and detectable genetic signatures, but we can expect to detect shared histories and shared DNA that cross the geo-political borders of our day. Especially with recent sharing of DNA, most ethnicity predictions at this level will have lower confidence.
This is a useful lens to keep in mind when examining your potentially conflicting ancestry results. Sub-regional level estimations are always less confident than predictions at the continental level. You would never expect to see an estimation that placed you as an Aboriginal Australian in one test, and Native American in another: these are deeply divergent branches of the human family. But when attempting estimations in the leafy twigs, you can expect to see some cross-over. Sweden in one test, Denmark in another? It’s possible. Germany in one test, Eastern Europe in another? Sure. This is the level of confident resolution that is the state-of-the-art of DNA ancestry prediction at this time, with the type of genetic markers and analysis used by companies today.
What to read next: Why confidence levels matter in DNA ethnicity
Shed light on your ethnicity results
Your ethnicity results can give you new insight about who you are and where your ancestors came from. Continue learning with our free ethnicity guide!

